As a reaction to the record high of fuel prices, the Portuguese government has updated the IVAucher program, to allow each citizen to recover 10 cents per each liter of fuel spent, up to a maximum of 5 EUR/month. This blog post is not going to discuss whether this is good way of spending the public budget, or if it is going to make a real impact in the lives of the people that manage to subscribe to this program. Instead, I want to focus on data.
Once you subscribe to the program as a consumer, you just need to fill the tank in one of the gas stations that subscribed the program, as businesses. The IVAucher website publishes a list of subscribed stations, which seems to be updated, from time to time. The list is published as a PDF, with 2746 records, ordered by “districto” and “concelho” administrative units.
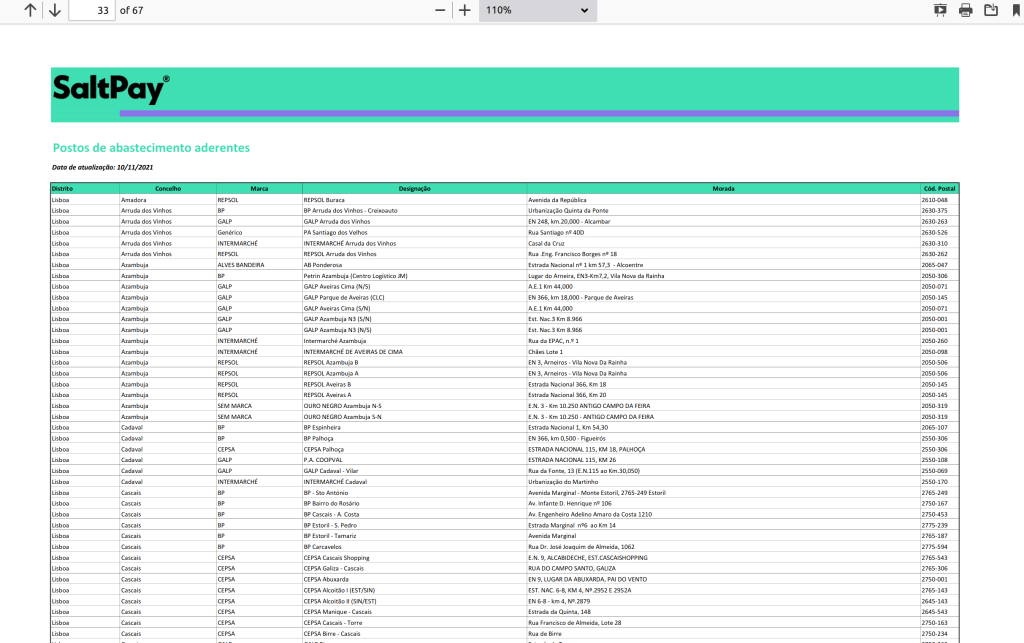
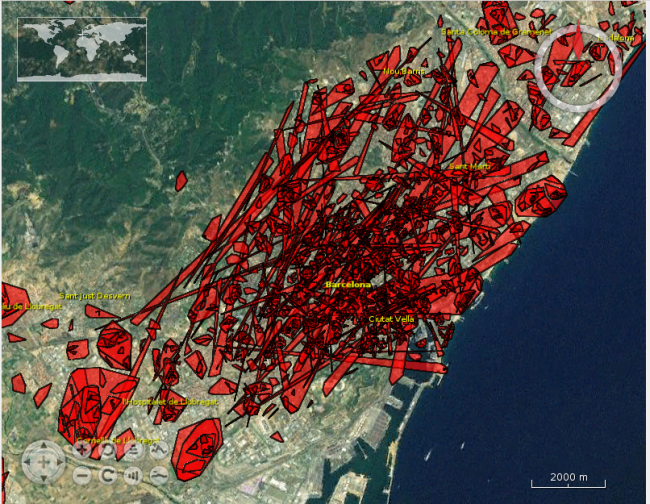
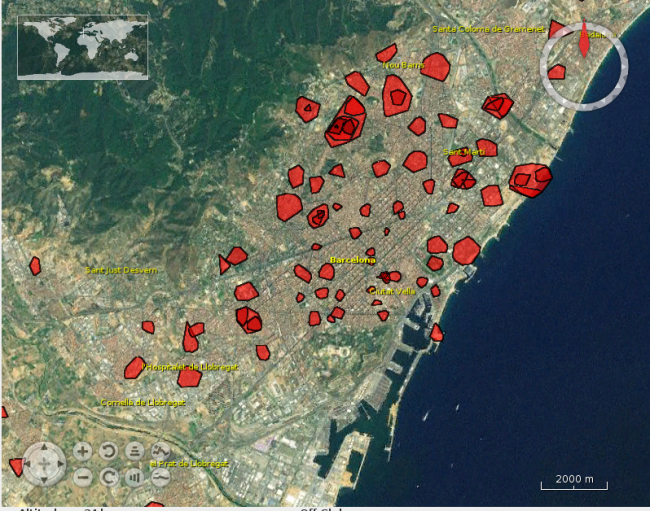
When I look for the stations around me, in the “concelho” of Lisbon, I found 67 records. In order to know where to go, I would literally need to go through each and check if I know the address or the name of the station. Lisbon is a big city, and I admit that there are lots of street names that I don’t know – and I don’t need to, because this is “why” we have maps. My first though was that this data belonged in a map, and my second though was that the data should be published in such a way that it would enable other people to create maps – and this is how this project was born.
In the five-star deployment scheme for Open Data, PDF is at the very bottom, and it is easy to understand why. There is so much you can do with a format, which is largely unstructured.
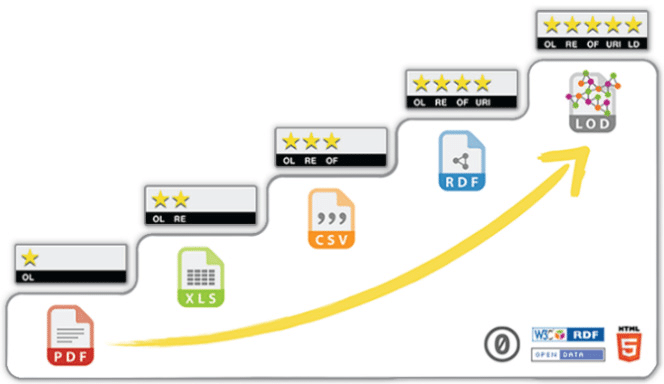
In order to be able to process these data, I had to transform it into a structured format, preferentially non proprietary, so I chosen CSV (3 stars). This was achieved using a combination of command-line processing tools (e.g.: pdftotext, sed and grep).
The next step was to publish these data, following the FAIR principles, so that it is Findable, Accessible, Interoperable and Reusable. In order to do that, I have chosen the OGC API Features standard, which allows to publish vector geospatial data on the web. This standard defines a RESTfull API with JSON encodings, which fits the expectations of modern web applications. I used a Python implementation of OGC API Features, called pygeoapi.
Before getting the data into pygeoapi, I had to georeference it. In order to do forward geocoding, I used the OpenCage API, and more specifically a Python client, which is one of the many supported SDKs. After tweaking the parameters, the results were quite good, and I was even able to georeference some incomplete addresses, something that was not possible using the Nominatum OSM API.
The next thing was to get the data into a format which supports geometry. The CSV was transformed into a GeoJSON using GDAL/ogr2ogr. I could have published it as a GeoJSON int pygeoapi, but indexing into a database adds support to more functionality, so I decided to store it in a MongoDB NoSQL data store. Everything was virtualized into docker containers, and orchestrated using this docker-compose file.
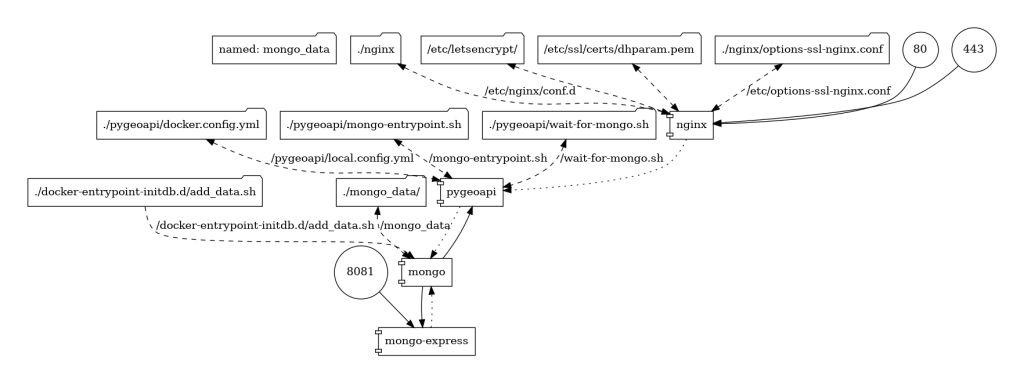
The application was deployed in AWS and the collection is available at this endpoint:
https://features.byteroad.net/collections/gas_stations
This means that anyone is able to consume this data and create their own maps, whether they are using QGIS, ArcGIS, JavaScript, Python, etc. All they need is an application which implements the OGC API Features standard.
I also created a map, using React.js and the Leaflet library. Although Leaflet does not support OGC API Features natively, I was able to fetch the data as GeoJSON, by following this approach.
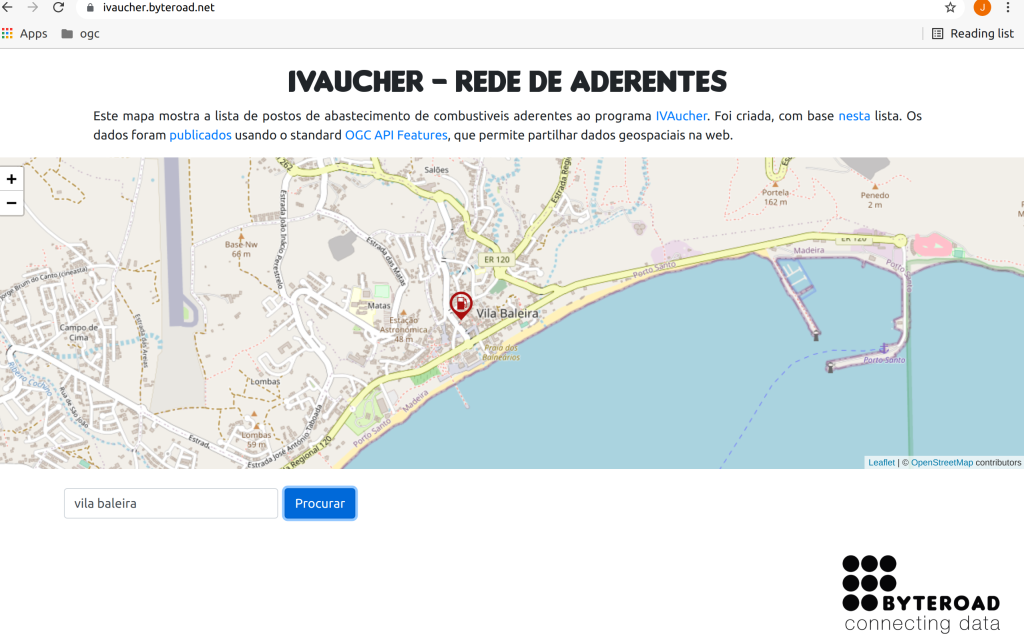
The resulting application is available here:
Now you can navigate through the map until you find you area of interest, or even type an address in the search box, to let the map fly to that location.
Hopefully, this application will make the user experience of the IVAucher program a bit easier, but it will also demonstrate the importance of using standards in order to leverage the use of geospatial information. Making data available on the web is good, but it is time that we move a step forward and question “how” we are making the data available, in order to ensure that its full potential is unlocked.